DynV
New member
- Joined
- Dec 22, 2017
- Messages
- 33
About the following translation on linear regression: What are the similarities and differences between homoscedasticity test of plain and Student residuals? Are they just 2 ways of doing the same technique of linear regression assumption verification, or are they 2 different techniques? If they're different, is there similarities in between, and if so which? The code is in R. There's text after the image. What's below is a translation of the lesson.
We can check the homogeneity of variances and normality using the same tools as for ANOVA.
Figure 3a shows that the variances are homogeneous. Although some residuals deviate from normality (3b), linear regression is appropriate.
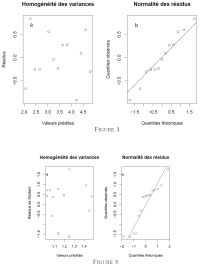
The diagnostic plots show that the variances are approximately homogeneous and that the residuals approximately follow a normal distribution (Fig. 8). Since we used Student residuals, we were able to confirm at the same time the absence of extreme values in fig. 8a).
We can check the homogeneity of variances and normality using the same tools as for ANOVA.
Code:
by(mfrow = c(1, 2), cex = 1.2)
##homogeneity of variance
plot(residuals(m1) ~ fitted(m1), ylab = "Residuals",
xlab = "Predicted values",
main = "Homogeneity of variances")
##normality
text("a", x= 2.3, y = 0.7, cex = 1.2)
qqnorm(residuals(m1), ylab = “Observed quantiles”,
xlab = "Theoretical quantiles",
main = "Normality of residues")
qqline(residuals(m1))
text("b", x= -1.4, y = 0.7, cex = 1.2)
Code:
##checks assumptions
by(mfrow = c(1, 2), cex = 1.2)
plot(rstudent(m.shocks) ~ fitted(m.shocks),
ylab = “Student Residuals”,
xlab = "Predicted values",
main = "Homogeneity of variances")
text(y = 1.5, x = 1.025, labels = "a")
qqnorm(rstudent(m.shocks), ylab = “Observed quantiles”,
xlab = "Theoretical quantiles",
main = "Normality of residues")
qqline(rstudent(m.shocks))
text(y = 1.5, x = -1.9, labels = "b")